Optogenetics:
Control of cytoskeletal dynamics via light mediated microtubule actin cross-linking
Control of cytoskeletal dynamics via light mediated microtubule actin cross-linking
|
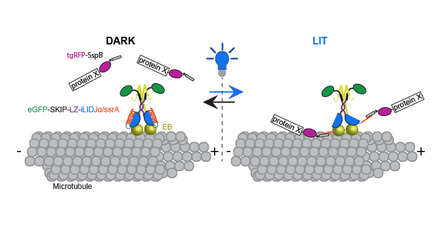
I developed, in collaboration with the Kuhlman lab at UNC, a novel optogenetic tool. This tool affords me the ability to spatially and temporally recruit proteins of interest to the microtubule plus end, allowing me to determine the spatial and temporal importance of these proteins throughout the cell cycle. I used this tool to investigate the role of temporal cross linking of the actin and microtubule networks. I am optimistic that the optogenetic tool that I developed can be transitioned into the organism, providing a means to investigate the spatial and temporal role of microtubule associated proteins during different stages of development at the sub cellular level within a developing animal.
|
Publication:
Adikes, R.C., Hallett, R.A., Saway, B.F., Kuhlman, B., Slep, K.C. (2018) Control of cytoskeletal dynamics via light mediated microtubule actin crosslinking. JCB 217(2) 779-793. DOI: 10.1083/jcb.201705190 Prepint on BioRxiv DOI: https://doi.org/10.1101/142414 |
Microtubule Polymerization:
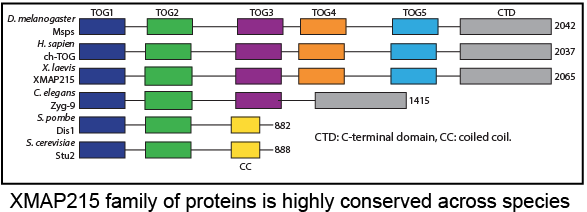
Structure Function Analysis of the C-Terminal Domain of the Drosophila XMAP215 Protein Family Member Minispindles
|
Publication:
Haase, K.P.*, Fox, J.C.*, Brynes, A.E.*, Adikes, R.C.*, Speed, S.K., Haase, J., Friedman, B., Cook, D.M., Bloom, K., Rusan, N.M., Slep, K.C. (2017) The S.cerevisiae XMAP215 family member Stu2 arranges its TOG domain array using a structurally distinct 15nm parallel coiled coil. *These authors contributed equally. Molecular Biology of the Cell. |
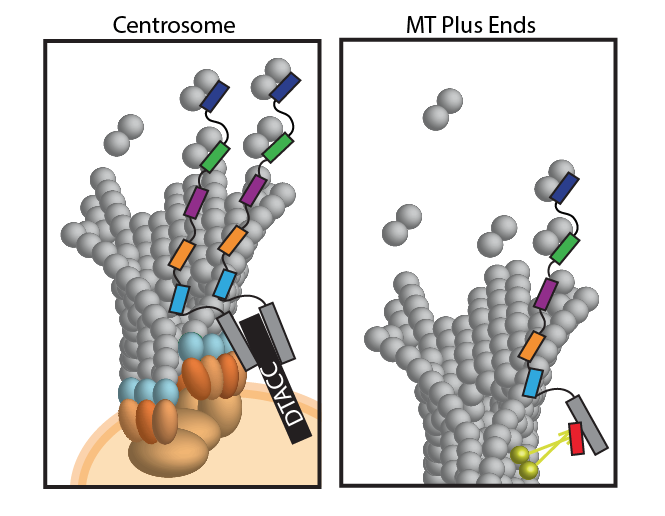
Microtubules (MTs) are cytoskeletal polymers responsible for multiple cytoplasmic activities: enabling intracellular transport, stabilizing the cell’s shape, and forming the mitotic spindle to separate chromosomes during mitosis. MT-associated proteins (MAPs) localize to MTs where they modulate MT dynamics through molecular and structural changes to the MTs. Members of a subgroup of MAPs that specifically localize to the MT plus end are referred to as +TIPs. A group of highly conserved +TIPs, known as the XMAP215 family, accelerate MT assembly and promote MT growth. XMAP215 family members promote polymerization via TOG (tumor over expressed gene) domains that bind to tubulin heterodimers. Studies of XMAP215 family members in multiple species (S. pombe, C. elegans, Xenopus egg extracts, Drosophila and HeLa cells) have shown that they play an essential role during interphase and mitosis, as their depletion leads to decreased MT growth rates and short spindles or defects in spindle architecture. Members of the XMAP215 family contain a varying number of N-terminal TOG domains, however, the structure and functional role of the C-terminal region remains unknown. My work in collaboration with three UNC undergraduates has shed light on the role of this C-terminal domain. |
FULBRIGHT RESEARCH
Non-Centrosomal Microtubule Nucleation and Organization during Mitosis
R.C.Adikes
- DAPI- Tubulin -Kif2a-
- DAPI- Tubulin -Kif2a-
Acentrosomal asters are small populations of microtubules that are nucleated on or around the condensed chromosomes at the beginning of mitosis. In the lab of Dr. Isabelle Vernos at the Center for Genomic Regulation in Barcelona, Spain I set out to determine if the mitotic kinesins and dynein played a role in nucleation, stabilization and/or organization of the acentrosomal asters. To address this question, I performed an RNAi screen of 10 different motors. Using a microtubule regrowth assay, I observed the nucleation and formation of acentrosomal asters through immunofluorescence and confocal time-lapse imaging. I found motors that seemed to affect microtubule nucleation or stabilization, microtubule polymerization, and microtubule organization. These experiments revealed a possible novel role for motor proteins during mitosis.
In addition to this cell work I learned how to harvest Xenopus laevis extracts, how to deplete key mitotic regulators from the extract and how to induce the formation of spindles in these extracts, in order to probe how key mitotic factors regulate microtubules dynamics and spindle formation.
In addition to this cell work I learned how to harvest Xenopus laevis extracts, how to deplete key mitotic regulators from the extract and how to induce the formation of spindles in these extracts, in order to probe how key mitotic factors regulate microtubules dynamics and spindle formation.
UNDERGRAD RESEARCH
Myosin 19 : Biochemical Analysis of a Mitochondrial Associated Myosin
|
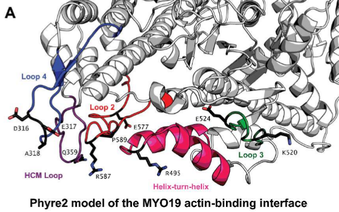
I worked with Dr. Omar Quintero at Mount Holyoke College and Penn State Hershey. The Quintero lab is interested in understanding the role of the actin based motor proteins, myosins, in mitochondrial movement. The lab previously discovered that myosin 19 (MYO19) co-localizes with mitochondria and thus could potentially play an important role in mitochondrial movement. Using Fluorescence Recovery After Photobleaching (FRAP) I determined that MYO19 exchanges slowly with other cellular pools indicating that MYO19 is tightly associated with the mitochondrial outer membrane. To further understand the mechanism of actin-based mitochondrial positioning in cells I probed the biochemical properties of this motor. In vitro studies of MYO19 revealed a slow rate of ATP turn over and suggested a medium affinity for actin, indicating that MYO19 is either a slow motor or an actin-based mitochondrial anchor. To determine if there were regions of MYO19 that could lead to these biochemical properties I performed sequence
alignments, charge determination and structural predictions of the motor domain. I was able to identify specific amino acids in the MYO19 motor that I hypothesize correlate to the observed biochemical data. Publications: Adikes, R.C., Unrath W.C., Yengo, C.M., and Quintero, O.A. Biochemical analysis of the myosin-XIX motor domain. Cytoskeleton 70(5), 281-9. Quintero, O.A., DiVito, M.M., Adikes, R.C., Kortan, M.B., Case, L.B., Lier, A.J., Panaretos, N.S., Slater S.Q., Rengarajan, M., Feliu M., Cheney R.E. (2009) Human myo19 is a novel myosin that associates with mitochondria. Current Biology 19, 2008-2013. |
Thin Filament Regulation
 Garland Publishing 1999
Garland Publishing 1999
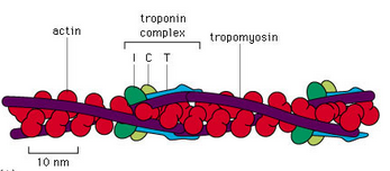
I worked with Dr. Ned Debold at UMass Amherst. The goal of this project was to combine the expression of mutant proteins with the functional assays to probe the structure/function relationship of proteins that are crucial to proper muscle and heart function. My experiments focused on determining the effects of a cardiomyopathy causing mutations in TnT, a subunit of the muscle regulatory protein troponin, on the regulation of actomyosin motility. As the lead person on this project I had the opportunity to teach other members of the lab basic DNA cloning, molecular biology techniques and epiflourescence microscopy.